R 2: Data Cleaning
Learning Objectives
As part of a research project you have been given some data that you are expected to clean yourself ready for analysis. The data is health report metrics taken from various London boroughs.
We have two goals for this project. The first is to clean the dataset, and the second is to make an aggregation on the cleaned data.
Note the data is randomly generated but is based on the kind of dataset would see in the real world; the deprivation data we are using is real.
Key takeaways from this worksheet:
How to use packages in R
Loading in datasets with different file extensions
Doing calculations across columns in a data frame
Using conditional operators (logic) to clean and prepare data
Finding and handling missing data
Transforming data through mutating joins
Exploring data through grouped aggregations
Project goals
The first goal is to clean your dataset so it looks like the data shown below. Take a few moments to have a look at the dataset. You can scroll to the right to view all the columns.
The second goal is to use that cleaned data to make a grouped aggregation of location vs average BMI and average IMD decile.
# A tibble: 8 × 4
location n avg_bmi avg_imd_decile
<chr> <int> <dbl> <dbl>
1 Hackney 364 23.9 2.82
2 Islington 361 23.9 3.75
3 Tower Hamlets 355 24.3 3.76
4 Southwark 343 24.0 3.88
5 Lambeth 384 24.2 4.04
6 Camden 385 23.8 5.52
7 Westminster 376 23.9 5.59
8 City of London 366 23.9 6.81Setup
- Download the zip file, by clicking the button below, and save on your computer. Try to save it somewhere where you can find it again, ideally not downloads!
Unzip the file. On a Mac you double click, on a Windows you right click and select extract all.
Open RStudio and open the
.Rprojfile. To open the project, go toFile > Open Project.Once your project is open, click on the
.Rscript or.Rmdfile you should see in the files panel on the right hand side of your screen. It is up to you which you use.In the files panel you should also see some data which are called
health_metrics.csvandIMD2019.xlsx. We will be loading these into R later.
For those that are interested R projects are a really handy way of organising your code. See a full explainer in the R for Data Science book.
Installing packages
For this project we are going to need some extra tools that are not included in the base version of R. Packages essentially are collections of functions people have written for everyone to use to make our lives easier, which is very helpful!
An important thing to note is that when installing a package you only need to do it once.
For this project there are two packages we recommend you install: tidyverse and janitor.
The tidyverse which is a set of tools designed to simplify data cleaning and wrangling tasks. It is a lot of different packages wrapped up in one. The two packages within the tidyverse we recommend using for this project are the dplyr package and the readxl package.
The janitor package contains several helpful utility functions that make common tasks easier. The documentation page has a lot of example use cases.
To install the packages we recommend you copy the code below and paste it into the console (bottom left panel in RStudio), then run it. For context, the Ncpus argument is there to speed up the install, which will take a few minutes.
install.packages("tidyverse", Ncpus = 6)
install.packages("janitor", Ncpus = 6)Loading packages
When using packages in R you need to load them each time you open R. We use the library() command for this, and the best practice is to do this at the top of your script/R markdown file.
At the top of your script/R markdown file use the library() command to load the tidyverse, readxl, and janitor packages.
Note that when loading the tidyverse set of tools it does not load everything it installed, but instead loads a handful of the packages it installed for us. The readxl package is not loaded, so we have to load that separately.
Hint: You will need to use the library function for each package you are loading!
Lets load the data so we can start cleaning it!
Load in both data files. Keep in mind that one file is a .csv file, and the other is a .xlsx file.
When you have them loaded, you should see them appear in the Environment panel (top right of RStudio).
Try taking a look at your data using the functions View(), str(), and summary() once the data is loaded.
Remember to assign your data a name to store it like we have done with variables. E.g. some_data <- read.csv("my_data.csv").
Remember that Excel files have different sheets. Which sheet in the IMD2019.xlsx file holds the data? If you are not sure try opening the file to check. How can you use that information to load the data from that sheet?
Top tip when loading data: move your cursor between the quotation marks (’’ or ““) and press tab. You should see a list of the files you can load! Press tab to select or click using your mouse.
A side note on the English indices of deprivation (IMD) data
The IMD2019.xlsx dataset is from the English indices of deprivation website 2019. Indices of deprivation (IMD) measure relative deprivation in small areas in England, where lower decile scores indicate a more deprived area.
Tidy up the column names of the deprivation (IMD) data
The column names in the IMD data are messy and challenging to work with in R. Look at the two examples below. Which has the clean R friendly column names? What is the difference between the examples?
[1] "LSOA code (2011)"
[2] "LSOA name (2011)"
[3] "Local Authority District code (2019)"
[4] "Local Authority District name (2019)"
[5] "Index of Multiple Deprivation (IMD) Rank"
[6] "Index of Multiple Deprivation (IMD) Decile"[1] "lsoa_code_2011"
[2] "lsoa_name_2011"
[3] "local_authority_district_code_2019"
[4] "local_authority_district_name_2019"
[5] "index_of_multiple_deprivation_imd_rank"
[6] "index_of_multiple_deprivation_imd_decile"To tidy up the column names of the IMD data we recommend using the janitor package and the clean_names() function. Examples can be found on this blog post or on in package documentation. This makes the column names more R friendly which will make working with the dataset easier.
The health dataset already has R friendly column names so we do not need to change that.
Some weird stuff is happening in the heights and weights columns…
We have our data loaded which is the first real hurdle, great stuff! However, this dataset is not perfect…
Have a look at the weight and heights column in the health metrics dataset. Do you notice anything strange?
It seems people in this data have given a variety of metrics for weight and height. For height, you’ll see a mixture of meters (e.g. 1.75), centimetres (e.g. 175), and feet (e.g. 5.74) responses. For weight you’ll see a mixture of stone (e.g. 9.02) or kilograms (e.g. 57.27).
We need to fix this in order to perform calculations on this data! Your goal here is to add new columns to the data where height is in meters, and weight is in kilograms.
The first 5 rows of the new columns you should have made are shown below. You can check this by viewing your cleaned dataset (try the View() function).
height_m weight_kg
1 1.8288 65.39683
2 1.7900 65.88000
3 1.8332 57.26984
4 1.7154 82.73000
5 1.4000 68.38095Tips to help cleaning the height and weight columns
This isn’t straightforward to fix so we have to make assumptions which are listed below.
if height is > 8 assume it is in centimetres
if height is > 4 and < 8 assume it is in feet
if weight is < 25 assume it is in stone
Below are some converting formulas to help you out:
centimetres to meters: \(centimetres \div 100\)
feet to meters: \(feet \times 0.3048\)
stone to kilograms: \(stone \times 6.35029318\)
We are going to need to use conditional operators to find and fix the incorrect data points. The table below contains the conditional operators you can use.
| Operator | Meaning |
|---|---|
> |
Greater than |
>= |
Greater than or equal to |
< |
Less than |
<= |
Less than or equal to |
== |
Equal to |
!= |
Not equal to |
!X |
NOT X |
X | Y |
X OR Y |
X & Y |
X AND Y |
X %in% Y |
is X in Y |
In R, the easiest way to conditionally fix data is to use the ifelse() function, or the case_when() function from dplyr. To add a new column the transform() function or the mutate() function from dplyr are easiest to use.
To help you get started you can copy and paste the scaffolding code below. You’ll need to fill in the spaces where the ... is. It is recommended you look up the ifelse() function online or in the help menu (bottom right, help tab).
health <- mutate(health,
height_m = ifelse(..., ...,
ifelse(... & ...,
..., ...)),
weight_kg = ifelse(weight < 25, ... / 0.1575, weight))Remember, to see what your data should look like see the output in Section 8.
Now that is fixed, we can calculate some health metrics!
Your goal here is to make new columns in your dataset that are calculations of BMI and waist-to-hip ratio.
If all goes well you should end up with the first 5 rows of the new columns you have made looking like below.
bmi whr
1 19.55349 0.7795927
2 20.56116 0.7811985
3 17.04144 0.9380042
4 28.11462 0.9094361
5 34.88824 0.7953994Do you remember from the first worksheet how we calculated BMI and waist-to-hip ratio? How do we do those calculations on columns in a data frame to make a new column?
Hint: when using the mutate function to add columns you can add multiple columns in a single mutate function. See the previous exercise where we added height_m and weight_kg.
Add some meaning with categorisation
Categorising your data is a helpful way of adding some meaning to the data you have. In the data we are using we might want to create a category that tells us if someone has high, low, or normal blood pressure, as well as health categories for BMI and WHR (waist-to-hip ratio). For example, it is easier to tell that someone has a healthy BMI from a text data point than from a number such as 22.45.
Your goal here is to categorise blood pressure, BMI risk, and WHR risk. The first 5 rows of the new columns you should have made are shown below.
blood_pressure bmi_risk whr_risk
1 normal Healthy Low risk
2 normal Healthy Low risk
3 normal Underweight High risk
4 normal Overweight High risk
5 normal Overweight Low riskBlood pressure can be categorised into high, low, and normal.
High blood pressure is when systolic pressure is 140 and over, and diastolic pressure is 90 and over.
Low blood pressure is when systolic pressure is 90 and under, and diastolic pressure is 60 and under
The waist-to-hip ratio (whr) metric is another measure of health that is designed to look for people at higher risk of conditions like heart disease or type 2 diabetes. See the table below to help you categorise whr scores. Remember, there are different risk scores associated to men and women!
| Health risk | Women | Men |
|---|---|---|
| low | 0.80 or lower | 0.95 or lower |
| moderate | 0.81-0.85 | 0.96-1.0 |
| high | 0.86 or higher | 1.0 or higher |
Can you find the BMI risk categories yourself?
Top tip for multiple categories
When you are making multiple categories things can get messy using the ifelse() function as you have to do something called nested ifelse statements. The ifelse() function works best on one or two categories.
We would recommend using the case_when() function from dplyr for multiple categories as it makes things much easier. Check out the examples in the documentation page to help you out! The 6th example shown will be particularly helpful.
Handling some of the missing (NA) values
You may have noticed by now that the dataset has some missing data, particularly in the age and occupation columns.
There are many ways of dealing with missing values, and it should be case by case how you approach it. Sometimes you need to remove them, sometimes you need replace them with the mean (known as imputation), sometimes you need to change the value from missing to something more useful.
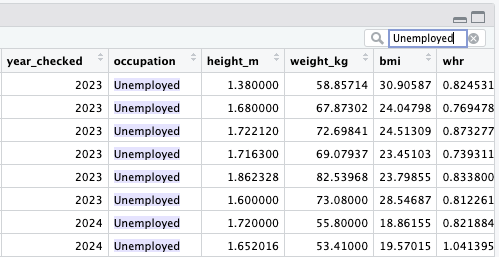
In this case, the missing values in the occupation column should be Unemployed rather than NA.
Using the techniques we have tried so far, change the NA values to Unemployed.
Hint: the is.na() function is really useful here.
To check if your conversion has worked, open the viewer by using the View() function or clicking on the dataset in the Environment window, and search Unemployed.
Congratulations, you’ve cleaned your dataset. Only one more preparation step to go!
Add the deprivation data to our dataset!
To add the deprivation data we loaded earlier from the IMD2019.xlsx file we need to use a technique called a join.
Your goal here is to use the join technique to bring both the datasets together. See the next section for help!
Once you have joined your two datasets we loaded earlier together your final dataset should look like the cleaned dataset below!
Another check you can do is see if your column names match those listed below in your final dataset. Particularly the lsoa_name_2011 through to index_of_multiple_deprivation_imd_decile columns. You can use the names() function to check your own data.
[1] "name"
[2] "sex"
[3] "age"
[4] "location"
[5] "lsoa_code"
[6] "waist"
[7] "hip"
[8] "systolic_pressure"
[9] "diastolic_pressure"
[10] "year_checked"
[11] "occupation"
[12] "height_m"
[13] "weight_kg"
[14] "bmi"
[15] "whr"
[16] "blood_pressure"
[17] "bmi_risk"
[18] "whr_risk"
[19] "local_auth_district_code"
[20] "imd_rank"
[21] "imd_decile"
[22] "lsoa_name_2011"
[23] "local_authority_district_code_2019"
[24] "local_authority_district_name_2019"
[25] "index_of_multiple_deprivation_imd_rank"
[26] "index_of_multiple_deprivation_imd_decile"Information on joins
A join is used to combine rows from two or more tables. There are four main types you are likely to need in R:
inner joinfinds matches between both data frames and drops everything elseleft joinincludes all of the data from the left data frame, and matches from the rightright joinincludes all of the data from the right data frame, and matches from the leftfull joinincludes all data from both data frames




In order to perform a join we need a column in each dataset, in this case health_metrics.csv and IMD2019.xlsx, to have matching data. Take a look now at the lsoa_code column in your health metrics data, and then the lsoa_code_2011 column in your IMD data.
What do you notice? Do you think we can use those to link up the datasets? If so, which join do you think should be used?
To perform a join using R, we can use the dplyr library. Below is some example code for you to look at and play around with.
library(dplyr)
person_info <- data.frame(
p_id = c(1,2,3,4,5,6),
Name = c("Andrew", "Chloe", "Antony", "Cleopatra", "Zoe", "Nathan")
)
food_info <- data.frame(
p_id = c(1, 4, 7),
Fav_Food = c("Pizza", "Pasta con il pesto alla Trapanese", "Egg fried rice")
)
person_food <- full_join(person_info, food_info,
by = join_by(p_id == p_id))
person_food p_id Name Fav_Food
1 1 Andrew Pizza
2 2 Chloe <NA>
3 3 Antony <NA>
4 4 Cleopatra Pasta con il pesto alla Trapanese
5 5 Zoe <NA>
6 6 Nathan <NA>
7 7 <NA> Egg fried riceWhat do you think would happen to the output if we used a inner join or a left join?
The dplyr documentation has a nice guide on joins in R. The R for Data Science book also has some useful information on this technique for further reading.
Lets aggregate!
Our final goal here is to do a little bit of exploratory analysis on our cleaned data! After looking at your data you might want to find out what the average BMI and average IMD decile is for each location. What location has the best or worst average BMI? Which location is the most or least deprived?
Your goal here is to have an output that looks like the aggregation we showed in the project summary, also shown below.
# A tibble: 8 × 4
location n avg_bmi avg_imd_decile
<chr> <int> <dbl> <dbl>
1 Hackney 364 23.9 2.82
2 Islington 361 23.9 3.75
3 Tower Hamlets 355 24.3 3.76
4 Southwark 343 24.0 3.88
5 Lambeth 384 24.2 4.04
6 Camden 385 23.8 5.52
7 Westminster 376 23.9 5.59
8 City of London 366 23.9 6.81But what is grouped aggregation? And how do I do it in R?
Grouped aggregation is the computation of summary statistics (such as the mean), giving a single output value from several variables. You can perform more complicated aggregations by grouping your data. This is similar to a pivot table in Excel. Have a look at the images below which break down what grouped aggregation is.
You can group your data by more than one group. This means when the data is split, more subsets are formed for all different possible splits.
The R for Data Science book has some helpful examples on how to do this in R as does the dplyr documentation.
Tip: you can use the arrange() function to order the output in a more meaningful way.
Save your cleaned dataset so you can use it later!
The final task is to save our cleaned dataset. We can then use that data for analysis and visualisation later!
Before we do that, it would be a good idea to tidy up some of the column names, and remove some of the columns we don’t need. The select() and rename() functions from the dplyr package will be really helpful for this.
To help you out we renamed the following columns:
index_of_multiple_deprivation_imd_ranktoimd_rankindex_of_multiple_deprivation_imd_deciletoimd_decilelocal_authority_district_code_2019tolocal_auth_district_code
And we removed the following columns:
lsoa_name_2011local_authority_district_name_2019height(as we have converted these to a new column)weight(as we have converted these to a new column)
Once you’ve renamed and selected the columns you need, write (save) your dataset to a .csv format. Save it in the same folder where your script/R markdown file and other data is stored, and make sure to give it a meaningful name such as health_data_cleaned for example.
Next steps
Great work on cleaning the dataset. When doing data work, most of our time is spent cleaning or preparing a dataset ready for the fun stuff, analysis and visualisation; depending on the data and project it can take up around 50-80% of your time!
Now you have cleaned the data, we will be using it for our visualisation and analysis worksheets!
Give us some feedback!
We are always looking to improve and iterate our workshops. Follow the link to give your feedback.
Solutions are available at the end of the survey.